Software Registry
XPLOR-NIH
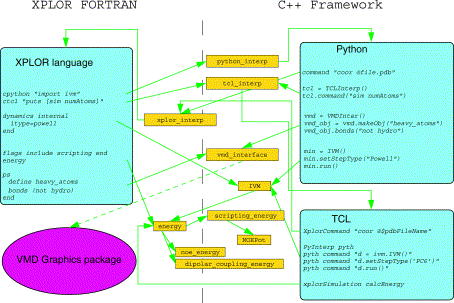
XPLOR-NIH is a structure determination program which builds on the X-PLOR v 3.851 program, including additional tools developed at the NIH. These tools include functionality for the following:
-
•3J couplings
-
•1J couplings
-
•13C shifts
-
•1H shifts
-
•T1/T2
-
•dipolar couplings
-
•radius of gyration
-
•CSA
-
•conformational database torsion angle potentials
-
•database base-base positioning potentials for DNA
-
•interface to the NMR graphics package VMD-XPLOR
-
•embedded Python and TCL interpreters.
Synopsis
Principal citations:
C.D. Schwieters, J.J. Kuszewski, and G.M. Clore, "Using Xplor-NIH for NMR molecular structure determination," Progr. NMR Spectroscopy 48, 47-62 (2006).
Documentation XPLOR-NIH
Related software




Keywords
Structure determination, molecular simulation, energy minimization